For more information about me and my research, visit my homepage:
PyNOT : scombine
Combining Spectra
PyNOT allows you to combine spectra of the same object directly from the terminal using the command pynot scombine.
The code automatically determines if the spectra are 1-dimensional or 2-dimensional and performs the combination accordlingly.
By default, the combination is performed assuming that the data are in units of flux (i.e., divided by exposure time).
If the exposure time and data units are correctly set in the FITS header, PyNOT will automatically detect if the data are
in flux units or in units of counts. Alternatively, you can set the command line option --scale to ensure
that the spectra are correctly scaled to the exposure time (or to the median flux in the spectra if exposure times are not in the headers.)
Make sure that the data have appropriately defined units! Otherwise you have to double check that the results have been correctly scaled to the total exposure time.
Combination methods
There are two methods available for the combination:
- Mean: will perform an inverse-variance weighted average. If the spectra have different wavelength coverage they will be interpolated onto a common wavelength grid covering the full extent of all the spectra unless using the option
- Median: will perform a median combination of all spectra. If the spectra have different wavelength coverage, only the overlapping parts of all the spectra will be median combined.
--trim in which case only the overlapping parts will be combined.
--method median (or -m for short).
Combination of 2D spectra
When combining 2-dimensional spectra, the code will try to identify the brightest object in the slit and align the exposures
to this position. This is useful when dealing with point sources that have been offset along the slit during the different exposures.
If you do not want to perform this object localisation and alignment, or if your object is extended or diffuse, you can turn this behavior
off by including the command line option -x --extended. When using the automatic image-alignment, there will be
regions on the sides where the data do not overlap due to the shifting of the two images. These regions can be trimmed away by using the
command line option --trim (or -t for short.)
For the object alignment to work properly, it is important that the dispersion axis is correctly identified. PyNOT will try to determine
this automatically, but if this fails, you can set the dispersion axis yourself using --axis:
- Horizontal spectra with dispersion along the x-axis:
--axis 1 - Vertical spectra with dispersion along the y-axis:
--axis 2
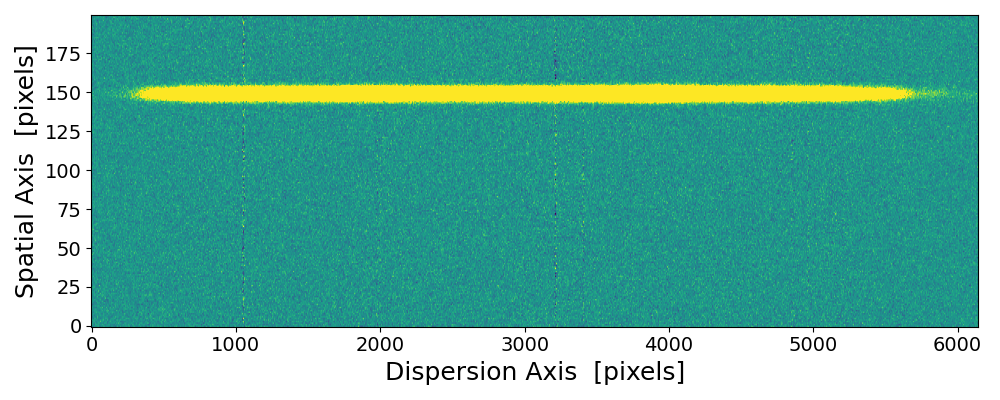
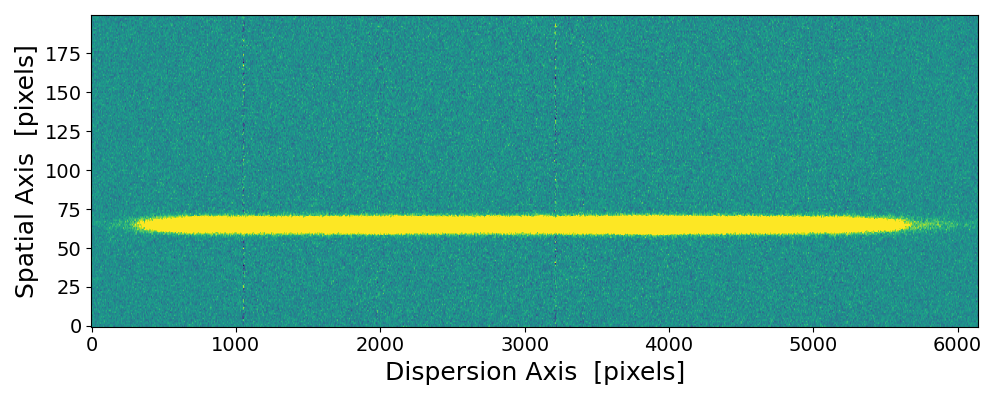
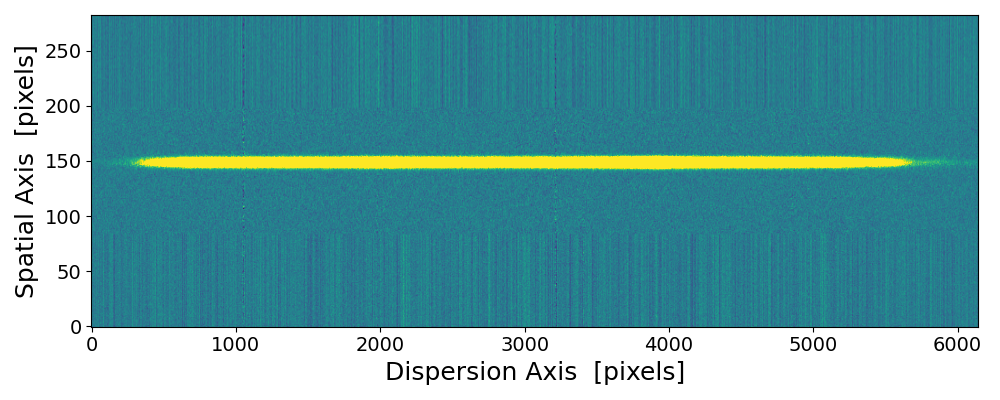
pynot scombine img1.fits img2.fits with default mean combination and object alignment.
The upper and lower edges are scrambled due to the image shift and interpolation applied when aligning the two images. These regions
are clipped if using --method median or the --trim option.
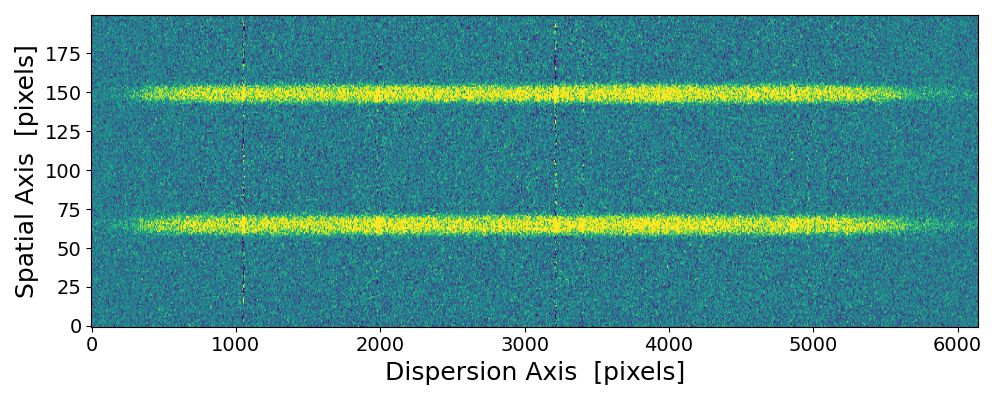
pynot scombine img1.fits img2.fits -x with default mean combination but ignoring the object offset.
Combination of 1D spectra
For the combination of 1-dimensional spectra, the options --axis and --extended are ignored.
However the other options have the same behaviour. By default, PyNOT works with FITS tables for the 1D spectra.
If you prefer to have your 1D spectrum saved as a multi-extension FITS (MEF) file with a 1D array in each extension for
flux, error, mask and sky (if present), then you can include the option --mef in the command line.
Example Syntax
pynot scombine input
Full example of command line syntax:
pynot scombine [-h] [-o OUTPUT] [-m {mean,median}] [-s] [-t] [--axis {1,2}] [-x] [--mef] input [input ...]
Overview of parameters
- input
- Input spectra
- --output (-o): ''
- Output filename (autogenerated if not given)
- --method (-m): 'mean'
- Combination method
- --scale (-s): False
- Scale spectra before combining
- --trim (-t): False
- Trim image regions that are not covered by all images (default when using method=median)
- --axis: 1
- Dispersion axis. 1: horizontal, 2: vertical (default: 1 for processed spectra)
- --extended (-x): False
- Set this option for 2D spectra of extended sources to turn off automatic localization
- --mef: True
- Set this option to save output as a multiextension FITS file instead of a FITS table.